Overview
EASI-FISH Pipeline
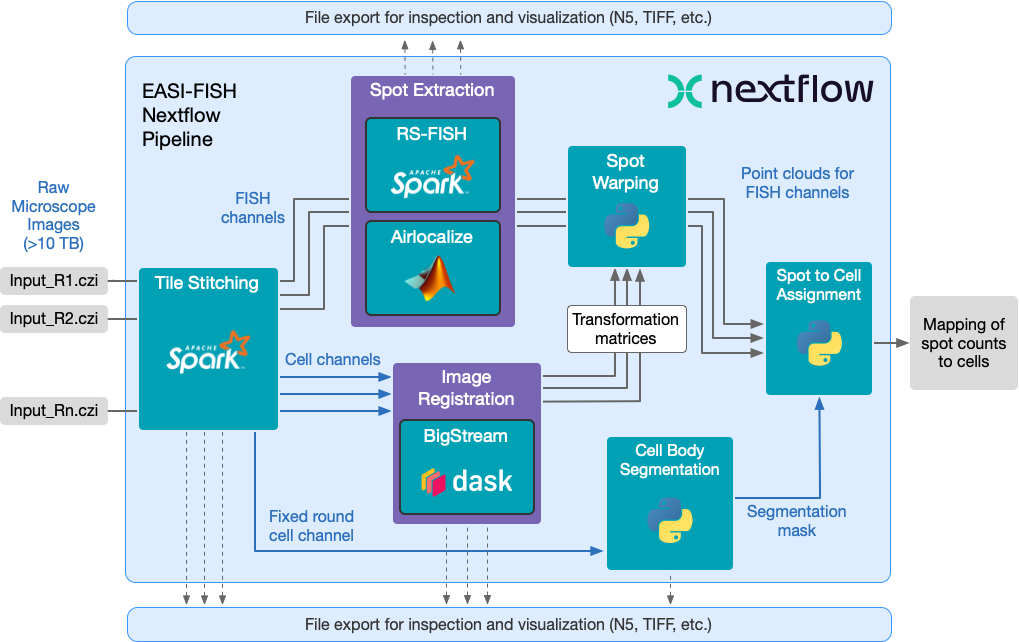
The purpose of this pipeline is to analyze imagery for spatial transcriptomics collected using EASI-FISH (Expansion-Assisted Iterative Fluorescence In Situ Hybridization). It includes automated image stitching, distributed multi-round image registration, cell segmentation, and distributed spot detection.
Modules
The pipeline includes the following modules:
- stitching - Spark-based distributed stitching pipeline
- spot_extraction - Spot detection using Airlocalize or RS-FISH
- segmentation - Cell segmentation using Starfinity
- registration - Round registration using the Bigstream distributed pipeline
- warp_spots - Warp detected spots to the round registration
- measure_intensities - Intensity measurement
- assign_spots - Mapping of spot counts to segmented cells
Some of these modules can be executed individually, using alternative main-*.nf scripts in the root project directory. You can also skip individual steps using the --skip parameter, as long as the processed data is placed in its expected location for any downstream steps.